41 match the phenotypes on the labels
Phenotype - Genome.gov Phenotype refers to an individual's observable traits, such as height, eye color and blood type. A person's phenotype is determined by both their genomic makeup (genotype) and environmental factors. Narration 00:00 … "Phenotype" simply refers to an observable trait. A cross-disorder dosage sensitivity map of the human genome 01.08.2022 · Finally, we discovered a strong relationship between the number of genes per segment and the number of associated phenotypes (Figure 2F; deletions, p = 1.81 × 10 −5; duplications, p = 1.62 × 10 −6; linear regression); pleiotropic segments (i.e., segments associated with multiple phenotypes) were also more likely to overlap strongly constrained genes …
quizlet.com › 586108722 › chapter-11-practiceChapter 11 Practice Problems Exam 4 Flashcards | Quizlet Offspring phenotypes If a parent has a heterozygous genotype for a trait, 1/2 of the time they will contribute a dominant allele to their offspring and 1/2 of the time they will contribute a recessive allele to their offspring. The phenotypes of the resulting offspring will be _____ dominant and _____ recessive.

Match the phenotypes on the labels
DailyMed - MECLIZINE HYDROCHLORIDE tablet Drug Label Information. Updated March 1, 2022 If you are a ... intermediate-, extensive-, and ultrarapid metabolizer phenotypes could contribute to large inter-individual variability in meclizine exposure. ... We anticipate reposting the images once we are able identify and filter out images that do not match the information provided in the ... › cell › fulltextA cross-disorder dosage sensitivity map of the human genome Aug 01, 2022 · This process yielded a total of 54 disease phenotypes, including 24 neurological, 28 non-neurological, and two general categories to capture broadly defined or nondescript phenotypes (Figure 1A; Table S2). Although imperfect, this strategy partitioned our dataset into 458,326 samples matching one or more primary disease phenotype (i.e ... Are mapped RefSeq, Ensembl, and UniProt IDs pointing to the … 17.08.2022 · Luckily the MANE labels have been integrated to BioMart so I was directly querying it. But it really doesn't cover many protein sequences, and some of the non-MANE ENSPs do match bidirectionally to RefSeq NP. Will see how the coverage goes. It looks like three-way mapping filters out more proteins, though the amount left is still acceptable. E.g.
Match the phenotypes on the labels. › p › 9534943Are mapped RefSeq, Ensembl, and UniProt IDs pointing to the ... Aug 17, 2022 · Luckily the MANE labels have been integrated to BioMart so I was directly querying it. But it really doesn't cover many protein sequences, and some of the non-MANE ENSPs do match bidirectionally to RefSeq NP. Will see how the coverage goes. It looks like three-way mapping filters out more proteins, though the amount left is still acceptable. E.g. scienceprimer.com › punnett-square-calculatorPunnett Square Calculator | Science Primer A Punnett Square for a tetrahybrid cross contains 256 boxes with 16 phenotypes and 81 genotypes. A third allele for any one of the traits increases the number of genotypes from 81 to 108. Given this complexity, Punnett Squares are not the best method for calculating genotype and phenotype ratios for crosses involving more than one trait. › gsea › docTable of Contents - GSEA | MSigDB Random phenotypes are created by shuffling the phenotype labels on the samples. For each random phenotype, GSEA ranks the genes and calculates the enrichment score for all gene sets. These enrichment scores are used to create a null distribution from which the significance of the actual enrichment score (for the actual expression data and gene ... Punnett Square Calculator | Science Primer A Punnett Square * shows the genotype * s two individuals can produce when crossed. To draw a square, write all possible allele * combinations one parent can contribute to its gametes across the top of a box and all possible allele combinations from the other parent down the left side. The allele combinations along the top and sides become labels for rows and columns within the …
GestaltMatcher facilitates rare disease matching using facial ... - Nature Disease genes such as PACS1 (gray) cause highly distinctive phenotypes but are ultra-rare, representing the limit of what current technology can achieve. The first new disease that was... II. INSTRUCTIONS: Match the letter of the definition that corresponds ... Guide Questions: 1. What are the possible phenotypes of the offspring based on the Punnett square? 2. Write the genotypic and phenotypic ratio of the offspring? ... Match the pictures in column A with its proper label in column B then; match the label in column B with its … correct description in column C. oh ano sino nakakita ng planet ... GEX_phenotype: Assignment of cells to phenotypes based on selected ... Description Adds a column to a VGM [ [2]] Seurat object containing cell phenotype assignments. Defaults for T and B cells are available. Marker sets are customizable as below Usage GEX_phenotype (seurat.object, cell.state.names, cell.state.markers, default) Arguments Value Returns the input Seurat object with an additional column Examples Re-identification of individuals in genomic datasets using public face ... specifically, these studies have shown that one can effectively match high-quality three-dimensional (3d) face maps of individuals with their associated low-noise sequencing data, leveraging known associations between phenotypes, such as eye color, and genotypes, which, for the purposes of this study, correspond to the variations in our genes …
Issues · genetics-statistics/GEMMA · GitHub Enforce failed for number of analyzed individuals equals 0. in src/param.cpp at line 2073 in ProcessCvtPhen. #262 opened on Apr 11 by zeroLB. 2 of 10 tasks. 1. Parsing input file '' failed in function AnalyzeBimbamGXE Support: use google groups instead. #261 opened on Feb 21 by MaxiEstravis. Issue with GEMMA-Wrapper. Punnett Square Calculator - Traits and Genes Calculator The Punnett square calculator provides you with an answer to that and many other questions. It comes as handy if you want to calculate the genotypic ratio, the phenotypic ratio, or if you're looking for a simple, ready-to-go, dominant and recessive traits chart. Moreover, our Punnet square maker allows you to calculate the probability that a ... Linear scoring - PLINK 2.0 14.08.2022 · The "range file" should have range labels in the first column, p-value lower bounds in the second column, and upper bounds in the third column, e.g. S1 0.00 0.01 S2 0.00 0.20 S3 0.10 0.50 (Lines with too few entries, or nonnumeric … Phenotype Matching | SpringerLink Self-referent phenotype matching is when animals learn and use their own phenotypes as referents for kin recognition (dubbed the "armpit effect" by Richard Dawkins in 1982). Self-matching provides the most accurate assessment of relatedness, as an animal's own cues better reflect its genotype than those of its close kin.
Table of Contents - GSEA | MSigDB Random phenotypes are created by shuffling the phenotype labels on the samples. For each random phenotype, GSEA ranks the genes and calculates the enrichment score for all gene sets. These enrichment scores are used to create a null distribution from which the significance of the actual enrichment score (for the actual expression data and gene set) is calculated. This is the …
Benchmarking atlas-level data integration in single-cell genomics - Nature 23.12.2021 · Single-cell atlases often include samples that span locations, laboratories and conditions, leading to complex, nested batch effects in data. Thus, joint analysis of …
Disaggregating Race/Ethnicity Data Categories: Criticisms, Dangers, And ... Constrained race/ethnicity options do not match how individuals self-identify and are racialized, leading to high rates of missing data and increasing selection of the "Some Other Race" option.
Brain-phenotype models fail for individuals who defy sample stereotypes ... Previous linear modelling work has relied on the assumptions that (1) a single brain network is associated with a given phenotype, with patterns of activity within that network varying across...
Guess her! : r/phenotypes 5.7K subscribers in the phenotypes community. a fun community to guess where someone is from based on how they look
BIO 105 SCIENCE OF BIOLOGY W/LAB - Essay Bros As more dark moths than light moths survived to reproduce, the ratio of light vs dark moths changed in favor of. the darker phenotype. PROCEDURE. Make a color copy of the Red Habitat (Figure 7) and the Blue Habitat (Figure 8), found on the next two pages. 1. Place 50 red and 50 blue beads into a 100 mL beaker.
Chapter 14 Mastering Biology Answers - Flashcards - StudyHippo.com Labels can be used once, more than once, or not at all. Click card to see the answer. answer. 1.) Probability that III-3 is a carrier (Rr): 2/3 2.) Probability that is a carrier III-4 is a carrier (Rr): 1/2 3.) Probability that IV-1 will be affected (rr): 1/12. Join StudyHippo to unlock the other answers.
Contrastive Learning of Single-Cell Phenotypic Representations for ... Phenotypes Profiling Cell images Cell representations Unsupervised learning Contrastive learning A. Perakis, A. Gorji and S. Jain—These authors contributed equally to the paper. Download conference paper PDF 1 Introduction An effective approach in the field of drug discovery is to relate treatments under development to existing ones.
Graphical View Legend - National Center for Biotechnology Information When the Pileup Display is set to "Match/Mismatch graph" (default), a pile-up graph displays above the alignments. Red markings in the graph denote mismatches while black markings denote gaps. ... When the Show Labels option is selected, labels for mate pairs are displayed. Figure 7.3.7. PCR duplicates, if indicated by the metadata in the ...
BIOLOGY H - Essay Help 1. In the circle below. sketch and label the two to three red blood cells and at least one white blood cell that you see using the hig… I need help with this. Match the terms shown below with the term that best fits. transcription factor linked to flowering in plants Responsible for formation of the Trade Winds and other wind patter…
CancerBERT: a cancer domain-specific language model for extracting ... We focused on 8 breast cancer phenotypes that describe the characteristics of breast cancer, including the Hormone receptor type, Hormone receptor status, Tumor size, Tumor site, Cancer grade, Histological type, Tumor laterality, and Cancer stage.
webTWAS: a resource for disease candidate susceptibility genes ... INTRODUCTION. First proposed by Gamazon et al. in 2015, the transcriptome-wide association study (TWAS) has emerged as a powerful method for investigating associations between genetic variants and disease or disease-related complex traits.TWAS utilizes a reference panel of genotype and expression quantitative trait data (such as GTEx ()) to fit a regression model predicting a target gene's ...
PLINK: Whole genome data analysis toolset - Harvard University Permute the dataset a large number of times, keeping LD between SNPs constant (i.e. permute phenotype labels) For each permuted dataset, repeat steps 2 to 4 above. Empirical p-value for set ( EMP1 ) is the number of times the permuted set-statistic exceeds the original one for that set.
Part A: Mix and Match Directions: Label the parts of Human Digestive ... Part A: Mix and Match Directions: Label the parts of Human Digestive System and pair it with its proper function by writing the letter beside each number S OF HUMAN DIGESTIVE SYSTEM FUNCTIONS A. Produces bile to break down fat. B. Connects the mouth to the stomach C. Stores bile and releases it when needed D. Absorbs nutrients from the digested food E.
Match the following terms and definitions. 1. replication ... - Weegy Match the following terms and definitions. 1. replication separation of DNA's two polynucleotide chains, which act as a template for a new chain 2. mutation when both genes are dominant and both are expressed in the phenotype 3. codominance when more than two alternatives for a gene exist 4. multiple alleles a copying mistake made during DNA replication 5. dihybrid cross the breeding of two ...
What is Cannabis Pheno-hunting? - Wikileaf If this is your goal, you know you'll need to grow out as many seeds as you have room for, find all the females from the batch, give them a label each female with a phenotype number ( #1, #2, #3…) to determine one from the other, and then flower them. It is very important to label each pheno with its number and strain labeled. Photo credit: Author
quizlet.com › 152624634 › chapter-11-masteringChapter 11 Mastering Biology Flashcards | Quizlet Drag the white labels to the white targets to identify the genotype of each F2 class. Remember that p (the pale mutant allele) and P (the wild-type allele) are incompletely dominant to each other. Consider the alleles for leaf shape next. Drag the blue labels to the blue targets to identify the genotype of each F2 class.
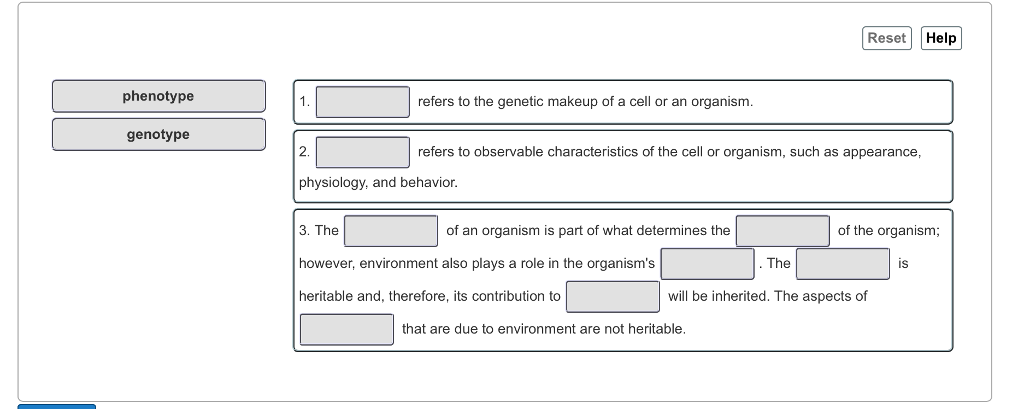












Post a Comment for "41 match the phenotypes on the labels"